DNA Profiling
Vocabulary
DNA polymerase – an enzyme that synthesizes
(builds) DNA from nucleotides.
DNA profiling – a forensic technique used to
identify individuals based on their DNA.
·
The
DNA profiles of suspects and evidence are compared to look for matches.
·
This
technique is also called DNA fingerprinting.
Gel electrophoresis – a technique used to separate
molecules (such as DNA) by size.
·
Charged
molecules move through a gel as an electric current is passed across it.
·
Smaller
molecules migrate through the porous gel farther than larger molecules.
Gene – a segment of DNA that gives
instructions for building a protein.
·
Other
than some small variations, the genes of different individuals are usually close
to identical and of the same length.
Mutation – a change in the DNA.
·
A
mutation in a gene may change the function of that gene. Mutations in non[1]coding regions
often have no effect on traits.
Non-coding region – a region of DNA that does not code
instructions to build a protein.
·
Non-coding
regions make up 99% of the DNA in the genome, while genes only account for
about 1% of the DNA.
·
Some
non-coding regions contain short tandem repeats (STRs).
Polymerase chain
reaction (PCR) – a
technique used to make multiple copies of a specific segment of DNA.
·
Primers
(short segments of DNA) bind to either side of a region of interest.
·
DNA
polymerase is then used to copy the DNA between the primers during a series of
heating and cooling cycles.
Primer – a short DNA sequence that provides
a starting point for DNA synthesis.
·
A
primer is designed to be complimentary to a specific region of DNA.
·
Two
primers that surround a region of interest in the genome can be used to amplify
(copy) that region of DNA.
Short tandem repeat
(STR) – a short
sequence of DNA, usually 2-7 nucleotides in length that repeat multiple times
in a row. STRs are also called microsatellites.
The number of repeats varies between different individuals.
·
Some
examples of STRs are AATG, TA, AGAT and GATA
Scientific Background:
DNA sequences consist of genes, or regions that code for
proteins, and non-coding regions that do not code for proteins. Short tandem
repeats (STRs) are short sequences of DNA, 2-7 base pairs long, which repeat
multiple times within the non-coding regions. During normal DNA replication,
STR regions are sometimes copied incorrectly, changing the number of STRs. As a
result, individuals have different numbers of STRs in the non-coding regions.
To compare the STR regions between different individuals, we
need a way to isolate and amplify those specific regions of DNA. Polymerase
chain reaction (PCR) is used to amplify specific regions of DNA.
Suppose you compare STR regions between individuals, like the
example shown at the right. In that case, DNA flanking (surrounding) STR
regions are the same between individuals, while the STR regions are variable in
length.
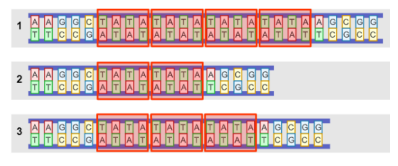
In PCR, a primer is a short
sequence of DNA used as a starting point for DNA synthesis. A pair of primers
are designed that are complimentary to the sequences on either side of the STR
region, as shown at right. While the primers in the Gizmo are only 3-6 base
pairs long, real primers contain 18-22 bases so that they are less likely to
bind to the wrong section of DNA.
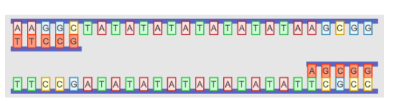
To begin PCR, a person’s DNA
is mixed with primers, DNA polymerase, and extra nucleotides. The mixture is
put through a series of warming and cooling cycles. In the first step of each
cycle, the mixture is heated, causing the DNA to denature, or separate. In the
next step, the DNA is cooled to allow the primers to bind to complimentary
sequences of DNA. In the third step, the temperature is raised again, which
allows DNA polymerase to begin replicating each strand, using the primer as a
starting point. Extension always occurs in the 3’ to 5’ direction of the DNA.
The top and bottom DNA strands point in opposite directions, so the products
look like this:

Notice the single-stranded
DNA on the left side of the top DNA copy and on the right side of the bottom
copy. This single stranded DNA is unstable and will eventually break down under
the heat cycles. The next two replication cycles are illustrated below. In each
cycle, the DNA strand is separated, primers are added to each side, and the DNA
is then copied from the primer to the end of the DNA. (Note, the primers are
shown in light pink below.)
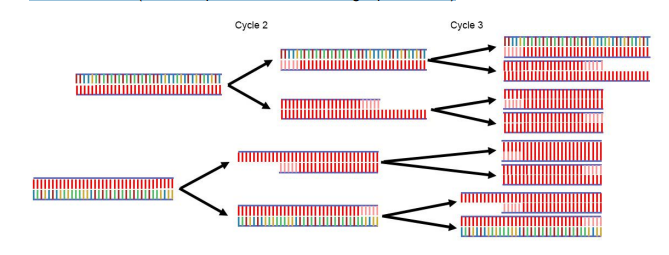
By the end of cycle 3, there
are 6 short segments of the region of interest and only two of the longer
segments. In the ensuing cycles, the number of shorter segments grows
exponentially until the shorter segments completely dominate the mixture.
Gel electrophoresis is a
technique used to separate molecules by size. Molecules, such as DNA, are
loaded into wells at the top of the gel. A negatively charged cathode is
attached to the top of the gel, and a positively charged anode is attached to
the bottom. DNA is negatively charged, so when an electric current is passed
through the gel, the DNA will migrate toward the anode at the bottom of the
gel.
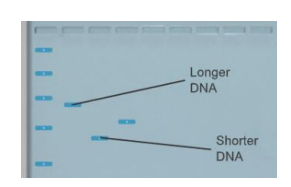
Inside the gel, the DNA
migrates through tiny pores. Longer DNA strands migrate more slowly than
shorter strands. Therefore, the longer DNA segments end up closer to the top of
the gel while the shorter segments end up towards the bottom, as shown in the
image at left. Fluorescent ethidium bromide (EtBr) is bonded to the DNA
molecules to allow the DNA to be seen.
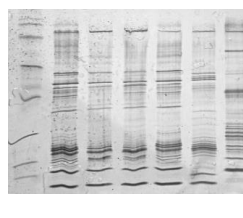
Using primers that surround
STR regions, PCR, and gel electrophoresis, a DNA profile (or DNA fingerprint)
is created (right). The profile is a series of bands representing each STR
region’s lengths. While two individuals may share some bands, it is extremely
unlikely that their entire DNA profiles will be identical.
Real World Connection:
DNA profiling, STR loci, and
CODIS DNA profiling can be used in criminal investigations to match a suspect
to evidence or it can be used to determine parentage by comparing a son or
daughter’s DNA to a potential parent.
First, DNA is extracted from
the cells in a sample. Then, several STR regions in each sample are amplified
using PCR. The FBI uses 13 different STR regions (loci) in their analysis. You
can find more information about each locus in the selected web resource below. The
STR regions in different individuals are variable, but each STR allele is
shared by about 5-20% of the population. The power of DNA profiling is in statistics.
If there is a 5% chance that two people will share each allele, there is a
1x10-17 chance that they will share 13 alleles. The 13 STR profile is compared
between individuals and a match is determined.
The FBI stores DNA profiles
in the Combined DNA Index System (CODIS). The system had been used to catch
criminals, but controversies have existed. The police are allowed to collect
DNA samples from people that were arrested, but not yet convicted, of a crime.
The CODIS system has also been used to find people that were relatives of those
already in the system. You can discuss the ethical implications of this with
your students.
The Innocence Project is an
organization that tests DNA evidence in criminal cases that occurred before DNA
profiling was available. They have exonerated hundreds of people who were
wrongly convicted of a crime. Darryl Hunt, the person used as an example in the
Student Exploration, is just one of the many people freed.